Detecting Cell Units (Neurons) and Their Activies
The past two years as my first postdoc project I started building a two photon microscope and virtual reality setup. The setup runs in a close loop so the animal behaviour can be mapped to its brain activity. To extract the neurons activities I ended up using CaImAn package in Matlab. To ease my pain and build a smooth pipeline for my application I build a script that calls CaImAn function in a certian order. The script starts with a popup that ask for user input for essential parameters and then ask for the image file. At the end, after the script finishes it will clean up the unncessary variables and save the rest for the user’s reference as a “.mat” file in the same directory.
CaImAn is a great work done by fine group of people. Please cite them and support them. For more details and learn about this package please visit their repository.
I have put together here my script and a striped down verstion of CaImAn with the hope that it might be of any assistance to anyone using CaImAn is a similar application.
Here is a test file for anyone to give it a try. One needs to only open and run “aaa_Extract_Calcium_Traces_04.m” which will promp for user input and the image file.
After the script rans, several figures will be popped out. I will briefly discuss the most important figures as a basic introduction.
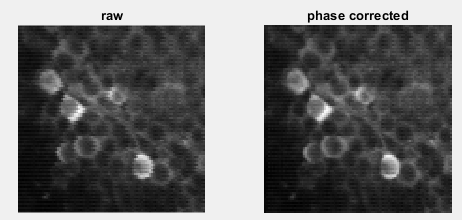
The first figure will be the result of phase correction. In systems that resonant scanners are used there will some shifts in pixels between the odd and even lines. So in the first stage of CaImAn, it will apply this correction to all the images. as show in the figure below, the corrected image has smoother and more organic edges.
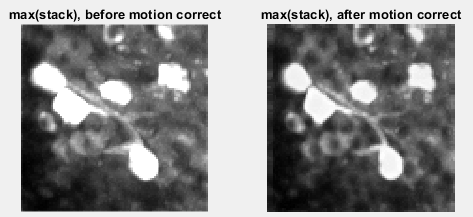
The next step is to account for the image drif during the course of acquisition. This is inevitable as the acquisition time can be very long. CaImAn tries to get a template from the first desired number of images and apply that to the whole stack. In the motion correction image, the maximum projection of the corrected stack looks more accurate in comparison to the raw data.
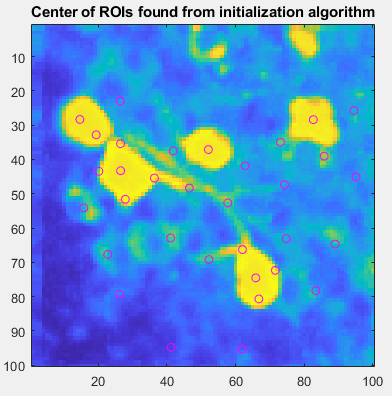
Now, CaImAn algorithm tried to find some starting points to initiated to detection method.
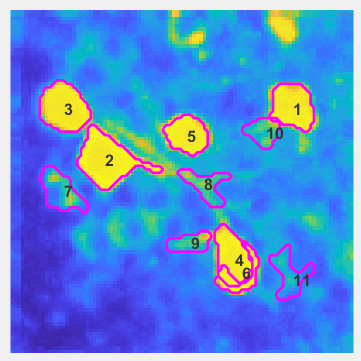
Next, a series of methods will kick in to find the active cells (units) in the image stack.
At the end, it will plot the traces of the detected units.
[Update 2022/08/24] I have uploaded a new script, “aaa_Extract_Calcium_Traces_05.m” that does some cleaning up for the starting number of selections. It does also clean up the detected cells that are not cells; region of interest is too small or too large compared to cell in the field of view.